Paula Ramos Giraldo, Søren Skovsen, Mayuresh Sardesai, Dinesh Bhosale, Maria Laura Cangiano, Chengsong Hu, Aida Bagheri Hamaneh, Jeffrey Barahona, Sandy Ramsey, Kadeghe Fue, Manuel Camacho, Fernando Oreja, Helen Boniface, Ramon Leon Gonzalez, Edgar Lobaton, Chris Reberg-Horton, Steven Mirsky.
Weeds are one of the biggest agricultural challenges; they compete with crops for available nutrients, water, and sunlight.
Myriad approaches, most notably RGB sensors, have been used to identify and map weeds using computer vision. For instance, a simple string weeder (aka a “weed eater”) paired with a camera can control very small weeds with unprotected growing points. As weeds get larger and more invasive, more expensive methods may be necessary, and some success has been had with robot-mounted lasers.
To date, however, no one has successfully integrated depth information simultaneously with object identification for the purposes of weed management. With 3D information, such as the kinds provided by OpenCV AI Kit with Depth (OAK-D), we can change control approaches based on where the growing point is and the overall hardiness of the plant.
Monitoring and mapping weed growth is one of the principal objectives of the Precision Sustainable Agriculture (PSA) team. The PSA team develops tools for monitoring the impact of climate, soil, and agricultural management on field crop production with the goal of improving farmer decision making.
We have generated a semi-automated and non-invasive method for the precise measurement of weed height and area covered under field conditions, using the new OpenCV camera, OAK-D, and the Intel Distribution of OpenVINO Toolkit. The project was well-received in the OpenCV AI Competition 2020 and won an honorable mention.
Here’s how we did it.
Hardware
First the field team went to the field and collected information from the transects, determined the type of weeds that had emerged, and measured weed height and ground cover.
Note: During the project, we had to change the camera used, as the OAK-D (BW1098OBC) was out of stock. Instead, we used a similar camera technology supported by the same software (Figure 1) in which components were not integrated into a single board. This version of the camera (BW1098FFC) has three FFC ports and an adjustable baseline, which allows for shorter minimum depth distances.
Set Up
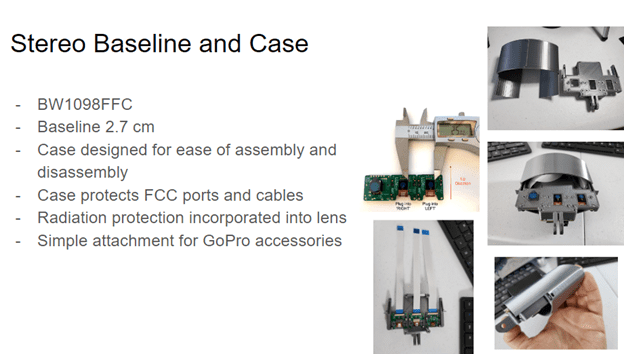

Materials list (Figure 2):
- Tablet (Lenovo P10 or M10 / or similar Android device)
- Telescopic monopod with strap
- OAK-D camera (BW1098FFC)
- Raspberry Pi 4 w/ micro SD card 32GB or bigger
- Power bank for camera and Raspberry Pi (2 outputs 10000mA)
- Cables
- Waist pack
- Tablet tripod mount adapter
- Mini ball head (#2)
- Lens cloth and brush.
Our objective was to create an easy-to-use, low-cost tool that will allow our national team of field scientists to quantify weed performance on-farm and on research stations.
We created five sensing systems with all of the components mentioned above and distributed them to our technology team for initial testing under diverse conditions in Texas, Maryland, Denmark, and North Carolina. Weed biomass was destructively collected to link images to weed performance.
Data Collection Methods

In order to create an optimal range of weed growth dynamics, we established field experiments at three locations including Texas, Maryland, and North Carolina (Figure 3). We included numerous plant species with different growth characteristics, including weeds and cover crops (CCs), to test OAK-D under realistic operating conditions.
The original plan was to run the DepthAI (Figure 4), collecting video while we walked slowly through the field.
Unfortunately, we had a synchronization issue in between the inference RGB camera and the stereo cameras: the RGB camera starts a few seconds before the mono cameras, which made it impossible to link packet sequence numbers.
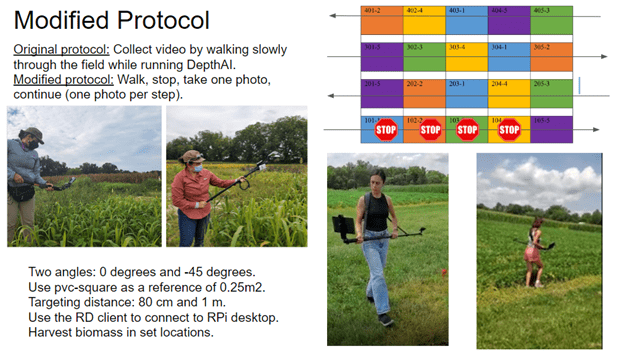
A revised approach required us to change the field data collection protocol. Instead of continually monitoring with video, we had to stop, take still images, and continue along the transect. We covered all transects, taking one still photo per step along the way.
Resulting Data
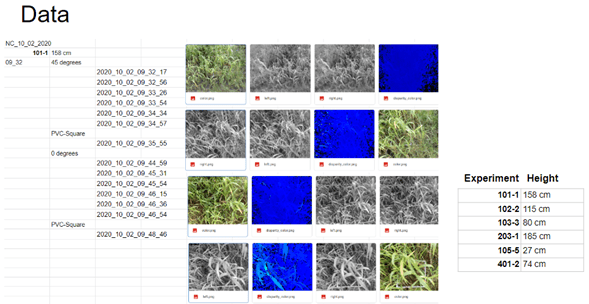
We used a modified Luxonis script to acquire raw depth information in a 16-bit NumPy array format, instead of a simple disparity map (Figure 5).
Due to image acquisition issues (auto exposure, auto white balance, and auto focus), we had to perform a raw database cleanup to run the biomass measurement detection models.
Semantic Segmentation
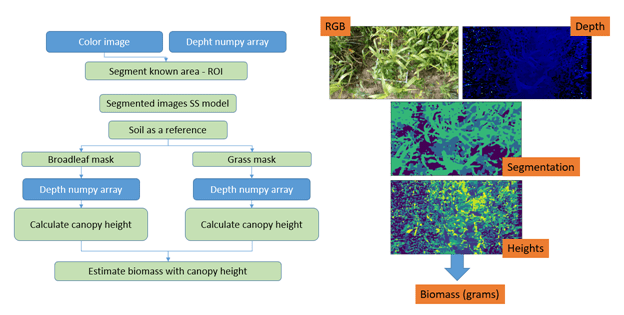
To predict weed composition, canopy images collected with the OAK-D camera were segmented into relevant categories of 1) soil, 2) grasses, and 3) broadleaf plants. This information was fused with depth measurements to predict not only the relative composition, but also the absolute values.
A DeepLab-v3+ model with a MobileNet-v3 backbone was implemented as a lightweight semantic segmentation model, suitable for the embedded environment. Compared to the commonly used backbone of Xception-65, this implementation has a reduced footprint with regard to memory and computing power, at the cost of lower accuracy.
Synthetic Data Creation
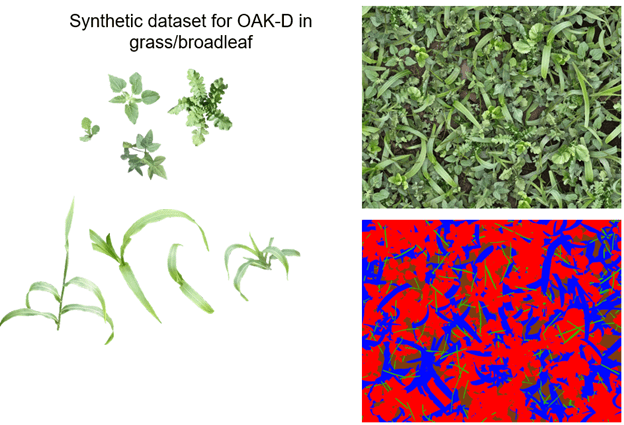
We trained the model in a supervised setting, requiring a dataset of images with accurate annotations for all pixels was required. Without regard to current research in one-shot learning, hundreds or thousands of labeled images are commonly needed to achieve sufficient generalization of the model.
Relying on the high similarity between plants within each species, a synthetic dataset was created to minimize labeling efforts. Following the principle of Skovsen et al. (2017) and Skovsen et al. (2019), as seen in Figure 6, 48 plant samples were digitally cut out of real images as demonstrated in the next figure and categorized as either a grass or broadleaf plant.
Model Testing
Because memory limitations reduce our segmentation model precision, we opted to continue development 1) using DepthAI on an Intel Neural Compute Stick and 2) processing the information directly in the host through the TensorFlow-lite library.
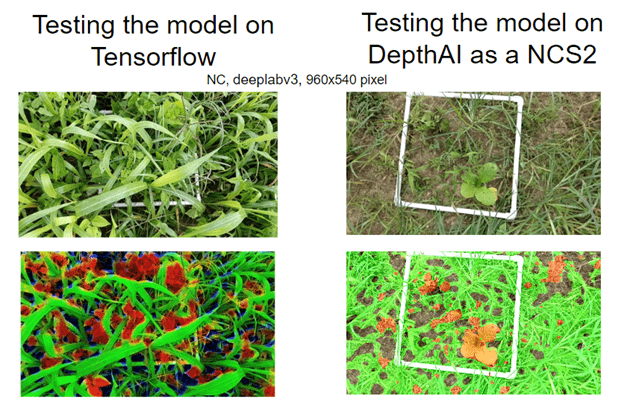
Once the image is acquired, it is processed by the semantic segmentation model, which detects grasses, broadleaves, and soil. The mask generated in the semantic segmentation will then be used with the depth maps also collected by the camera (which estimate heights of plants detected) to estimate biomass. Figure 8.
Development Process
We started by finding an RMS error in depth measurement, then we attempted to run the models in NCS2 mode. DepthAI was able to run the models but in our testing at the time only worked well on smaller models, returning segmentation faults on the larger ones.
We modified the data collection script to collect raw depth data into NumPy arrays to make and volume analysis less intensive. These images were then used to calculate volume based on a region of interest extracted using a white PVC pipe of a known area as a reference. The same images were used for static image analysis by conducting a 3D reconstruction and recovering the point cloud of the data.
Further experiments will involve improving the ROI-based approach as well as using the point cloud for volume estimation. Real-time processing did not work well because the segmentation quality decreased when we used smaller model sizes. As a result, we have decided to use non-real-time approaches for now. You can find our code on Github.
Results
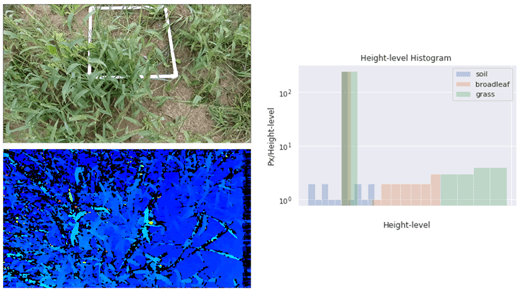
With our strategy, we are able to use the camera to collect plant heights. Using the histograms of heights generated from the host, we reduced the dimensions with Principal component analysis, took the first two components, and used them to make a multi-linear regression model to find the estimated biomass.
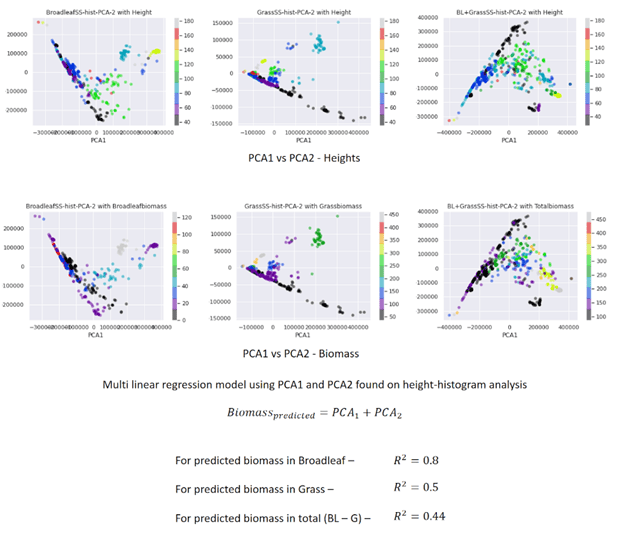
Determination coefficients between 0.5 and 0.8 show the potential of the OAK-D camera to measure the biomass of the weeds present in our fields.
Moving forward we intend to improve the accuracy of depth measurement using the new features of the DepthAI platform and address video data collection issues to achieve real-time analysis.
Team & Acknowledgements
We are part of the Precision Sustainable Agriculture team, and participated in the OpenCV Spatial AI Competition with our proposal “Weed monitoring”. In this competition, we collaborated with Aarhus University, North Carolina State University, USDA-ARS, and Texas A&M University.
Our thanks to Luxonis for their constant and timely help with all of our bugs and issues.
Our thanks to Julian Cardona for his 3D camera case design.
This research is based upon work that is supported by the National Institute of Food and Agriculture, U.S. Department of Agriculture under award number 2019-68012-29818, Enhancing the Sustainability of US Cropping Systems through Cover Crops and an Innovative Information and Technology Network.








5K+ Learners
Join Free VLM Bootcamp3 Hours of Learning